Ministère de l’Éducation Nationale
UNIVERSITÉ MONTPELLIER II
SCIENCES ET TECHNIQUES DU LANGUEDOC
IUP
GÉNIE MATHÉMATIQUE ET INFORMATIQUE
TRAINING COURSE SUMMARY
for
CIRAD (French Agricultural Research Centre for International Development),
tree crops department (CP),
at
the CBGP (Center for Biology and Management of Populations)
from october 2001 to january 2002
by
Alexandre Alapetite
Training course directors :
Informatics tools development for
identification of animal and vegetal populations
using information from the
DNA of a small group of individuals.
|
Université Montpellier II
IUP Génie Mathématiques et Informatique
Case courrier 025
Place Eugène Bataillon
34095 Montpellier cedex 5
Secrétariat : +33.4.67.14.49.52
Fax : +33.4.67.14.49.53
|
|
Centre de Biologie et de Gestion des Populations
CBGP Campus International de Baillarguet
CS 30 016
34988 Montferrier-sur-lez
Tél : +33.4.99.62.33.00
Fax : +33.4.99.62.33.45
|
|
CIRAD dép Cultures Pérennes
programme Cocotier
Avenue Agropolis
34398 Montpellier cedex 5
Tél : +33.4.67.61.58.00
Fax : +33.4.67.61.59.86
|
Acknowledgements
I wish to thank people without who this training course would not have been possible.
Special thanks to Sylvain Piry, Luc Baudouin and Chantal Hamelin that have built this project.
Thanks to Jean-Marie Cornuet, always there when a theoretic problem has to be solved...
Generally, I am proud of the confidence that everybody has shown, for technical choices has an example.
I don’t forget the CBGP team, especially Sandrine, Florent, Karine...
Summary
My training course subject was to build a component integrating a data structure for genetic of populations.
A program using this component has been developed for assigning
individuals or samples of individuals of unknown origin to populations.
Development has been made for Windows and Linux thanks to Delphi/Kylix.
When this training course ended, the program was working fine.
The main innovations where the assignation of samples of individuals (more than one),
and support of n-ploid individuals (individuals that have 1, 2 or more copies of their DNA information).
The program, and especially the data structure component, is foundation of a 3 years project (ATP).
While this project, modifications will be done, and new methods and ideas will be added.
The ability of evolving easily has been a priority.
The quality of the graphic interface did not suffer from that, and is easy to use, even for non-statisticians.

Table of Contents (Index)
Required notions
Introduction
A training course is done the first semester of the third year of
IUP GMI (MS in Mathematics and Informatics) of the university of Montpellier II.
I did it at the CBGP of Montpellier for the tree crops department of CIRAD.
This training course took place in a 3 years collaboration plan (Programmed Thematic Action)(ATP) between CIRAD, INRA, CBGP...
This is detailed later.
Heart of problem: assignation
Biologists from INRA.CBGP and CIRAD.CP build statistics on genetic information (genotypes)
taken from samples that can be pools of vegetables or animals.
Thanks to genetic characteristics of studied individuals,
it is possible to generate statistical characteristics of full samples.
The aim of assignation is to find out what are the samples that are close to reference populations that are well known.
Same calculation can be made on isolated individual.
Biology notions
Each individual, animal or vegetal, has copies of the information of its DNA.
Individuals that have 1, 2, 3,... copies are called respectively haploid, diploid, triploid,...
A locus is a short portion of chromosome (fragment of DNA), holding a code written with A, C, T, G bases.
So a polyploid individual may have different versions of the information of a locus.
The different versions are called alleles.
Chromosomes contain infinity of locus.
But all loci are not technically usable or interesting.
Most of the time, 10 to 20 loci are used; chosen for their polymorphism (different versions of the same locus are known).
Statistics are built with alleles found on chosen loci,
for several individuals of a sample to obtain alleles apparition frequency.
This is called allele frequencies (for a sample or a population).
Allele frequencies can characterise a population.
This characterisation will be more precise if many individuals and loci have been studied.
In practice, biologists do not have many individuals by sample (5-30), so allele frequencies are imprecise.
Statistics
The problem treated while this training course is test samples assignation to reference populations:
- There is a database containing allele frequencies for well known reference populations.
- Allele frequencies are calculated for a test sample made by several individuals.
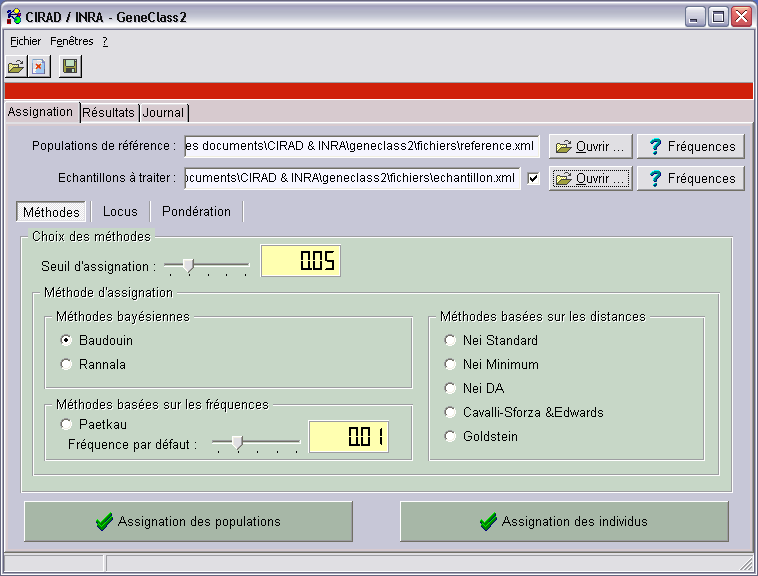
- Different statistic methods (Cornuet, Paetkau, Goldstein, or another Bayesian method)
can tell what reference populations are closest to the sample.
Integration
CIRAD is a French scientific organization specializing in agricultural research for the tropics and subtropics of the world.
Its mission is to contribute to rural development in the countries of these regions through research,
experiments, training, and dissemination of scientific and technical information.
Its work covers agricultural, veterinary, forestry, and food sciences.
CIRAD’s international cooperation activities cover more than 90 countries in Africa, Asia,
the Pacific region, Latin America, and Europe.
CIRAD’s researchers are posted in 50 countries;
they work with national research organizations or provide technical support in development projects.
CIRAD’s research centres are located in Montpellier, Greater Paris, and Corsica in France,
and in the French overseas territories of French Guiana, French Polynesia, Guadeloupe,
Martinique, Mayotte, New Caledonia, and Réunion.
CIRAD employs 1800 people, including 900 senior staff.
Its budget amounts to 1 billion French francs (€ 152 million).
CIRAD’s 7 departments operate 28 research programmes:

Coconut plays an important role in tropical economies and farming systems,
primarily in Asia, the Pacific and in coastal and island areas.
The crop is above all grown on smallholdings, partly as a food crop-for its water,
meat and sap-whilst also providing growers with a regular income from copra production.
In recent years, copra has been faced with stiff competition from other tropical and even temperate oil crops,
and the sector could well die out in the least competitive areas.
Against this backdrop, the Coconut Programme has chosen to focus its research on improving crop productivity and producer incomes,
on integrated control of lethal decay diseases and on diversifying the outlets for coconut.
Objectives:
- To increase coconut productivity, particularly on smallholdings.
- To restore the competitiveness of copra, the main source of vegetable oil in producing countries and the principal world source of lauric oil.
- To keep coconut in the traditional growing zones, for its food, economic and cultural value, and to develop alternative outlets for smallholders.
- To prevent the risk of coconut disappearing from regions affected by lethal decay diseases
Luc Baudouin, member of this programme, is my training course director at CIRAD.
His job is to select coconuts. For that, he has to be able to identify coconuts genetically.
He has made a coconut database with 600 individuals, for about 100 populations.
National Institute for Agronomic Research.
The CBGP (a joint research unit INRA/IRD/CIRAD/AGRO.M) aims at understanding the processes regulating biological
populations important in Agriculture, Environment and Human health.

The applied objective is to contribute to improve the strategies of control (especially biological control)
of pests and to identify the strategies of endangered natural populations conservation.
The estimation of gene flows between populations is favoured,
to forecast the dissemination of specific genes, deliberately introduced or selected.
The characterization of the genetic systems involved, the analysis of their interactions,
the determination of their adaptative value in various environmental contexts, experimentally estimated when possible,
are the knowledge required in order to control the population dynamics and manage the pests.
At the same time, the impact of environmental factors in the target environment-population systems is considered.
This approach lies on the coupling of the populations’ demographic analysis with that of the surrounding physical environment.
Modelling is a key-tool, both to orientate the research hypotheses and to integrate the various analyses’ scales.
Modelling at the scale of spontaneous or cultivated ecosystems allows the elaboration of decision-enabling tools
and to develop alternative methods of control and protection.

Populations studied often had important demographic variations that disturb normal statistic analyses.
Team 2 is concentrated on developing genetic methods that care about demographic variations.
Team 2 is directed by Jean-Marie Cornuet.
Sylvain Piry, has been my training course director.
He is working on informatics solutions for biology.
ATP
My training course has been financed by a 3 years ATP (Programmed Thematic Action).
This ATP, witch is cooperation between INRA, GEVES and different departments of CIRAD
has been proposed by Luc Baudouin and has started on 09/10/2000.
The total budget is 124K€.
Personal integration
I have been welcomed into team 2 of CBGP.
Thanks to the warm environment, my integration has been fast and I have quickly discovered the subject.
I had to recover my old biology notions to get in touch with the theory.
Starting point
Needs and existing solutions
Some programs had been made to solve calculations about populations genetic.
Unfortunately, each one uses its own file format.
They are sometime hard to use and some functionalities lacks.
Most of the time, they can only work for diploids.
There is a waste of time because several programs are needed to do one job.
I would be good to have only one program with:
- Easy-to-use and efficient data structure.
- Generalisation of assignation algorithms.
- Everything has to work under Windows and Linux.
- Specification of a file format that can store all the needed information.
- French and English versions.
- Test, implementation and validation of some assignation algorithms.
- ...
Results
CLX components, an intermediate and reusable result
Components are solutions to reuse peaces of programs easily and many times.
CLX (cross-platform component library) is a components library that can be used on Windows and Linux.
TPGDSStructure, main data structure
This is the earth of my job. It can read and write to standard files,
and allows manipulations of individuals and populations with their genetic characteristics.
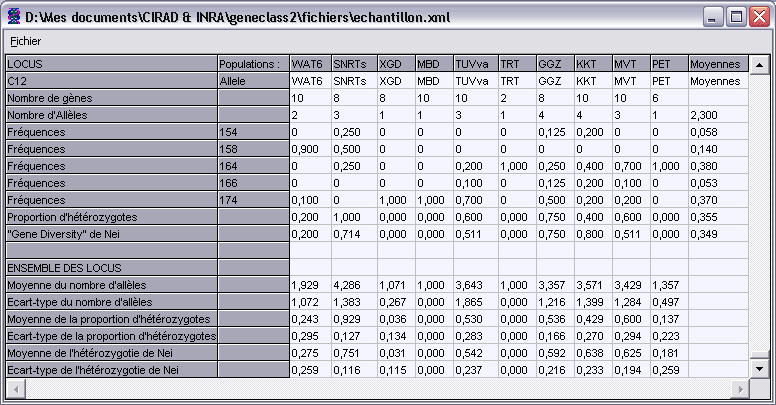
TFrequencyDisplay, graphical report
This graphical component is a window that shows statistics from a TPGDSStructure data structure.
For each population and for each locus, it displays the number of genes founded,
the number of alleles and their frequency, and other things.
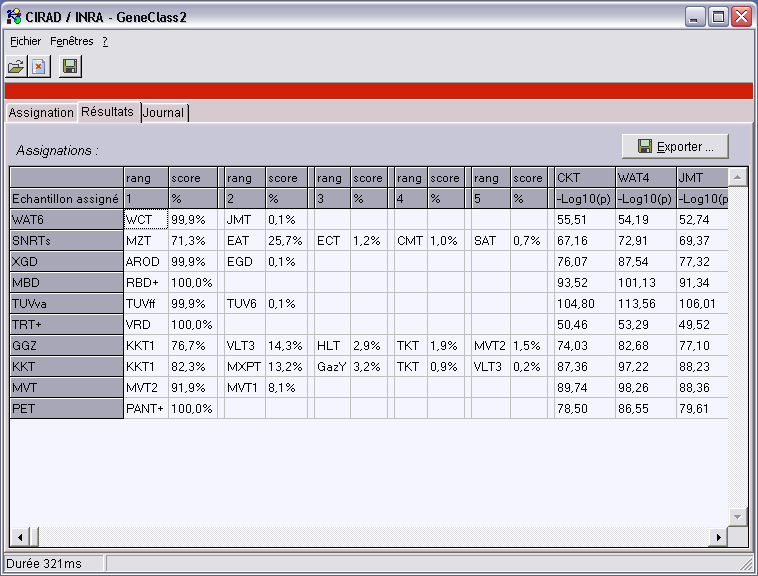
GeneClass 2, assignation program
GeneClass 2 is a software that uses several TPGDSStructure components and allows n-ploid
individuals or samples assignations to reference populations.
It can work on very big samples (tested example 500.000 fishes for a biologist in Quebec) with millions of individuals.
That could not be done with previous solutions.
Several scientists all over the world (Australia, Quebec, England, France...) look forward to its validation
and release into public domain.
Conclusion
This training course has been a big work. More than 14.000 lines would have been necessary for GeneClass 2 delphi project.
I have been pleased of chosen tools (Delphi, Kylix, XML, Lex & Yacc, ...).
Needs and objectives have been satisfied.
GeneClass 2 is been validated by clustered tests (lots of tests on many computers).
The statistic assignation method from Luc Baudouin has been published.
This training course has been positive for both sides, and I hope to find such an exiting job.
- Update from November 2003:
- Geneclass 2 is available on the CBGP’s softwares page.
- Update from October 2004:
- Publication of GENECLASS2: A Software for Genetic Assignment and First-Generation Migrant Detection by the CBGP team.
Alexandre Alapetite